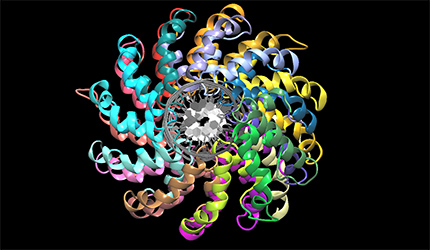
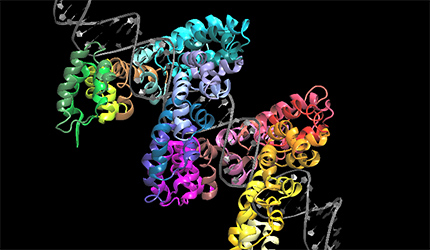
We work on the structure-function relationship of plant metallothioneins, structural analysis of plant TOLL-interleukin I like resistance genes and TALE protein derived from the plant pathogen Xanthomonas. We study plant G-proteins involved in signal transduction using modeling and synchrotron X-ray diffraction and scattering methods. We perform protein structure prediction using molecular modeling, homology modeling and various other computational tools. A circular dichroism (CD), FPLC and HPLC equipment are routinely used for structure prediction and protein purification.
The group working on proteins is interested in factors affecting the biological activity, viscoelastic properties of proteins, conformational multiplicity, conformational search on loop regions in proteins and tolerance of protein function to sequence variation.
Understanding Protein Function through Structure and Dynamics
We seek to design conditions under which proteins are encouraged to sample conformations that serve a specific purpose. This includes pH and ionic strength modulation, as well as point mutations introduced at locations that are prone to inducing mechanical effects. We utilize atomistic and coarse-grained simulations supplemented by theoretical developments to cover the pico-millisecond time scales, and nano-micrometer length scales. We have developed network-based models to study the basic features contributing to structure/function relationships in these systems. These include:
- Elastic network models in 1-D (GNM) and 3-D (ANM). (ANM server is located at http://midst.sabanciuniv.edu/anm)
- Optimal paths along the networks to identify key locations participating in their proper functioning. (available as a server under Services link at http://midst.sabanciuniv.edu)
- System identification based on perturbation/response scanning (PRS) to study the near-equilibrium dynamics. (PRS server is located at http://midst.sabanciuniv.edu/prs)
- Prediction of relaxation times contributing to equilibrium fluctuations.
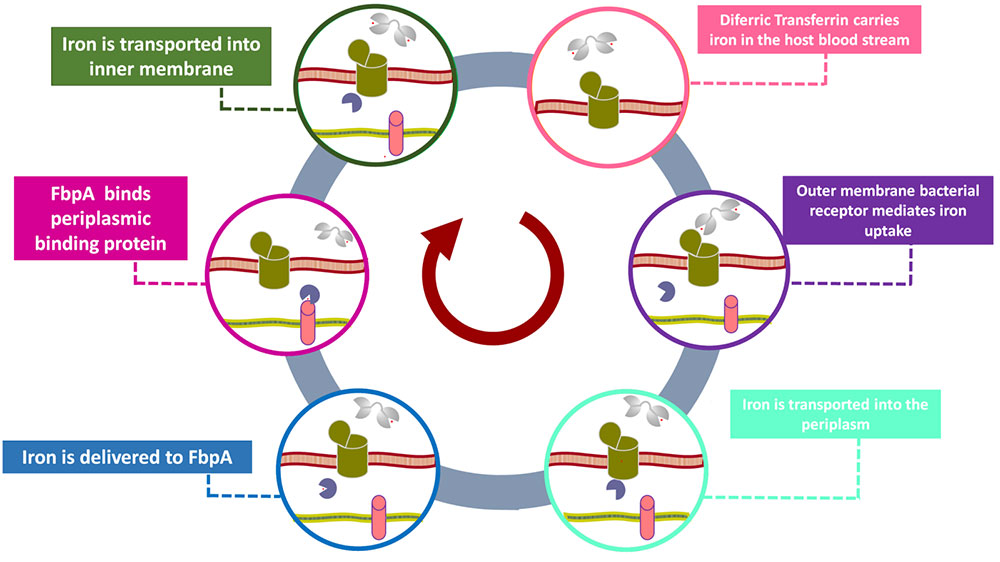
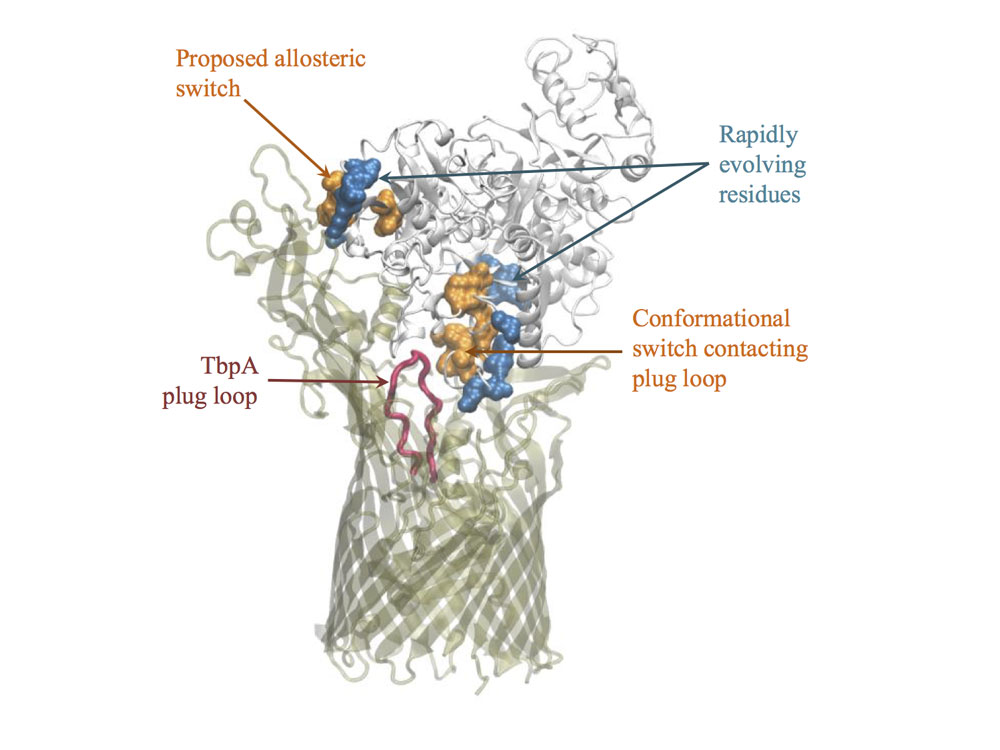
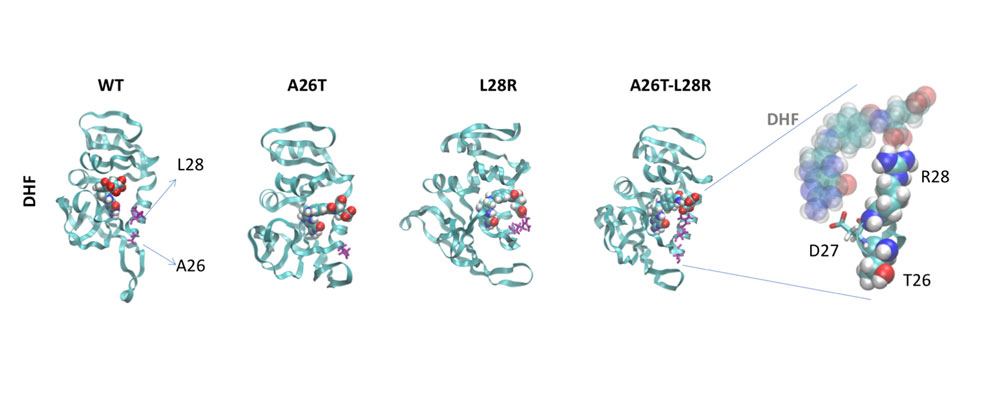
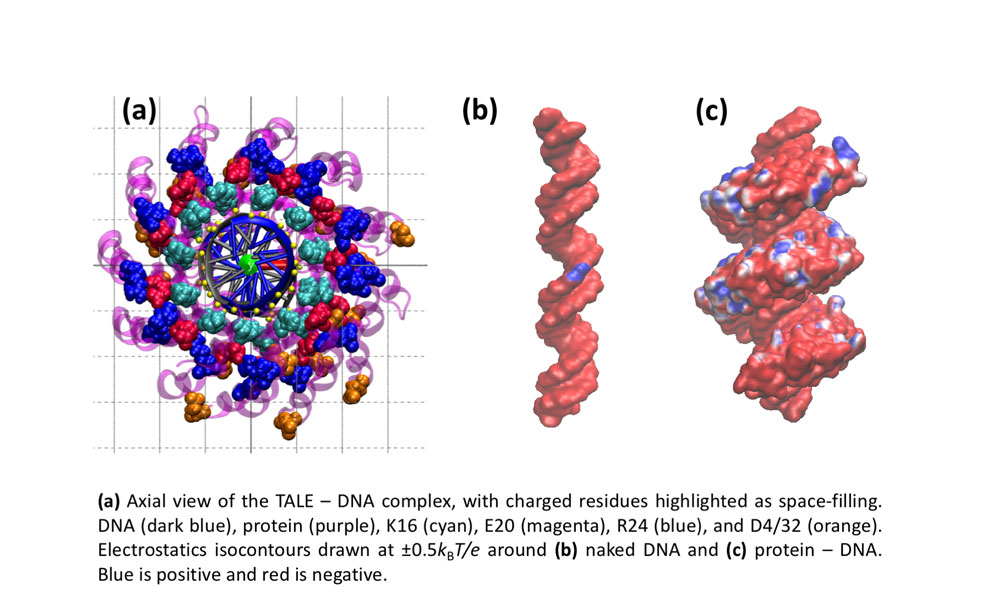
Some problems of interest are, iron piracy from humans by bacteria, molecular mechanisms that lead to mutants conferring resistance to bacteria in competitive inhibition, DNA binding properties of TALE proteins to design better TALENs, tetramerization of proteins with key roles in human cancer.
Structural Proteomics and Data driven Modeling
New developments in mass spectrometry have opened up the possibility to infer structural information of proteins and large protein complexes on a proteome-wide scale. We are developing methods and web-services that use chemical cross-link data to drive different molecular modeling simulations. Furthermore, proteomics data from limited proteolysis coupled to targeted mass-spectrometry experiments provides us information on subtle and global conformational changes on proteins. We use the data to establish the activity of enzymes, identify targets for small molecules or determine protein stabilities; all on a proteome-wide scale.
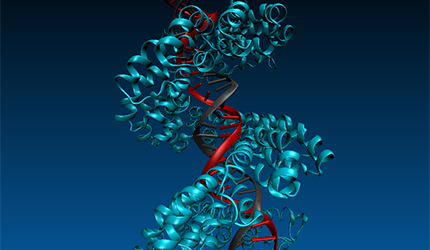
Further InformationPictured: Molecular dynamics studies on TALE DNA binding proteins from plant pathogenic Xanthomonas bacteria. Information derived from naturally occurring TALE proteins is used to design TALEN proteins which are biotechnological tools that mutate desired gene loci in complex genomes such as the human genome. We are investigating the DNA binding properties of TALE proteins to design better TALENs.